Decoding the Mitochondrial Unfolded Protein Response
Summary of a Nature paper from Goethe University Frankfurt revealing the previously unknown mechanisms of the mitochondrial unfolded protein response.
Written by Joris Frenz; Proteomics and Cancer Cell Signaling PhD researcher at the German Cancer Research Center, Heidelberg
Summary of the paper: Sutandy, F.X.R., Gößner, I., Tascher, G. et al. A cytosolic surveillance mechanism activates the mitochondrial UPR. Nature. PMID: 37286597.
In the complex world of cellular biology, maintaining protein homeostasis, or proteostasis, is crucial for proper cell function and survival. As proteins perform a large variety of essential tasks within cells, any disruption in their folding and function can lead to cellular stress and dysfunction. Importantly, mitochondria have their own quality control system called the mitochondrial unfolded protein response (UPRmt).
In the article “A cytosolic surveillance mechanism activates the mitochondrial UPR”, Sutandy et al. uncover the fascinating realm of the UPRmt, exploring its signaling mechanisms and its role in maintaining mitochondrial proteostasis.
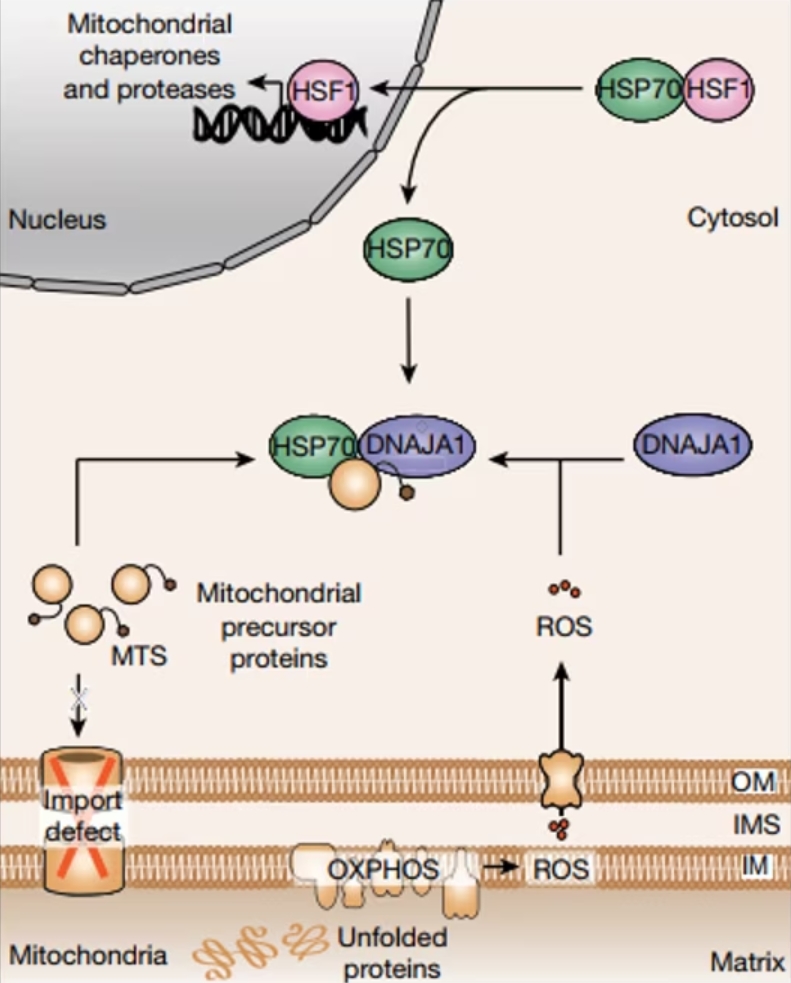
Figure 1: Schematic representing the cytosolic UPRmt surveillance mechanism resulting in transcriptional UPRmt activation. Figure from Sutandy et at., 2023. PMID: 37286597. Open access.
What Is the Mitochondrial Unfolded Protein Response?
The UPRmt is a cellular process that safeguards mitochondria from proteotoxic damage by activating a dedicated transcriptional response in the nucleus. When encountering mitochondrial misfolding stress (MMS) through the accumulation of misfolded or unfolded proteins, the UPRmt is triggered to restore proteostasis within these vital organelles. While the importance of the UPRmt is well established, the mechanisms through which mitochondria signal MMS to the nucleus have remained elusive.
Mitochondrial Reactive Oxygen Species (mtROS):
As a first step, the researchers induced UPRmt and performed subsequent RNAseq, which showed a significant increase in transcription for proteins involved in the response to oxidative stress. Further, mitochondrial reactive oxygen species (mtROS) were found to be increased in cells undergoing MMS. Hence, while mitochondria are known to produce ROS as byproducts of their normal functioning, oxidative stress appears to play a crucial role in the UPRmt. During MMS, the production of mtROS increased and they subsequently acted as signaling molecules, diffusing into the cytosol to convey the stress signal. Further, it was shown that knockdown of the HSP40 protein DNAJA1 inhibited the activation of mitochondrial protease LONP1 by inducing oxidative stress through GTPP. This finding indicates the crucial role of DNAJA1 in UPRmt induction through mtROS.
Accumulation of Mitochondrial Protein Precursors (c-mtProt):
Alongside mtROS, the study highlights the role of accumulated mitochondrial protein precursors in the cytosol (c-mtProt) as a critical signal for UPRmt activation. Mitochondrial proteins are synthesized in the cytosol as precursors and then imported into the mitochondria, where they undergo processing to achieve their mature functional form. With MMS in yeast, import defects can lead to the accumulation of these precursors in the cytosol, signaling the need for proteostatic intervention. A similar mechanism is now shown in human cells; preventing c-mtProt accumulation in the nucleus by translational inhibition with cycloheximide significantly reduced UPR signaling. A similar effect can also be shown by the depletion of mitochondrial biogenesis factor NRF1, indicating that this signaling pathway is selective for mitochondrial protein precursors.
The Link between Mitochondrial and Cytosolic Proteostasis:
Both mtROS and c-mtProt signals integrate to activate the UPRmt. Released mtROS oxidize the cytosolic HSP40 protein DNAJA1, which enhances the recruitment of cytosolic HSP70 to c-mtProt. Consequently, HSP70 releases HSF1, a transcription factor that translocates to the nucleus and activates the transcription of UPRmt genes. Importantly, no relocalization of DNAJA1 upon oxidation is observed. This highly orchestrated cytosolic surveillance mechanism ensures the restoration of proteostasis within mitochondria.
The discovery of this signaling mechanism reveals an intriguing connection between mitochondrial and cytosolic proteostasis networks. While a canonical UPRmt was not previously described in yeast, it was observed that c-mtProt accumulation could trigger the remodeling of cytosolic proteostasis in yeast and other organisms. In humans, the UPRmt serves as a more complex cytosolic surveillance system that integrates signals to activate the UPRmt. This link between mitochondrial and cytosolic proteostasis opens up new avenues for understanding diseases where breakdowns in cytosolic proteostasis are associated with mitochondrial dysfunction.
Implications and Future Directions:
The findings presented in the study have broad implications for our understanding of cellular proteostasis and its regulation. Further research into the UPRmt may unravel the intricacies of how cells maintain the delicate balance of protein folding and function, which is vital for their survival. Exploring the signaling pathways involved in UPRmt activation could provide insights into potential therapeutic strategies for diseases associated with mitochondrial and cytosolic proteostasis dysregulation.
Moreover, understanding the UPRmt and its connections to cytosolic proteostasis sheds light on the cellular processes involved in aging and age-related diseases. Mitochondrial dysfunction and impaired proteostasis are hallmarks of aging, and deciphering the mechanisms that maintain mitochondrial and cytosolic protein homeostasis may offer potential avenues for interventions to promote healthy aging.
The integration of signals from mitochondrial reactive oxygen species (mtROS) and the accumulation of mitochondrial protein precursors (c-mtProt) highlights the interconnectedness of mitochondrial and cytosolic proteostasis networks. These findings pave the way for future investigations into cellular proteostasis and its relevance to human health and disease.
PTG products used in this publication:
|
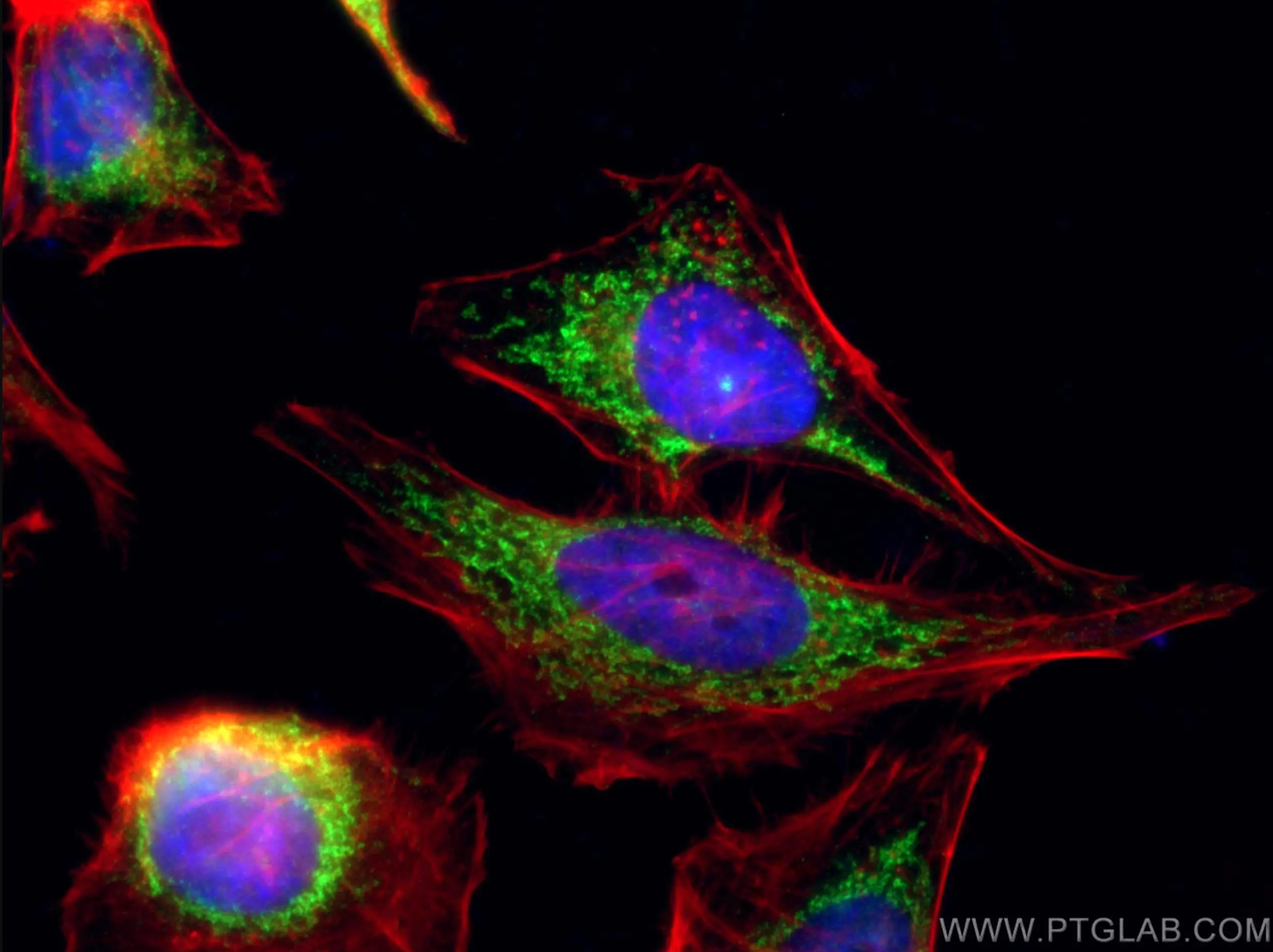
Featured Product COX5B Polyclonal Antibody (11418-2-AP)
Immunofluorescent analysis of (4% PFA) fixed HeLa cells using 11418-2-AP (COX5B antibody), at dilution of 1:100 and CoraLite®488-Conjugated AffiniPure Goat Anti-Rabbit IgG(H+L). Phalloidine stains the F-actin. |
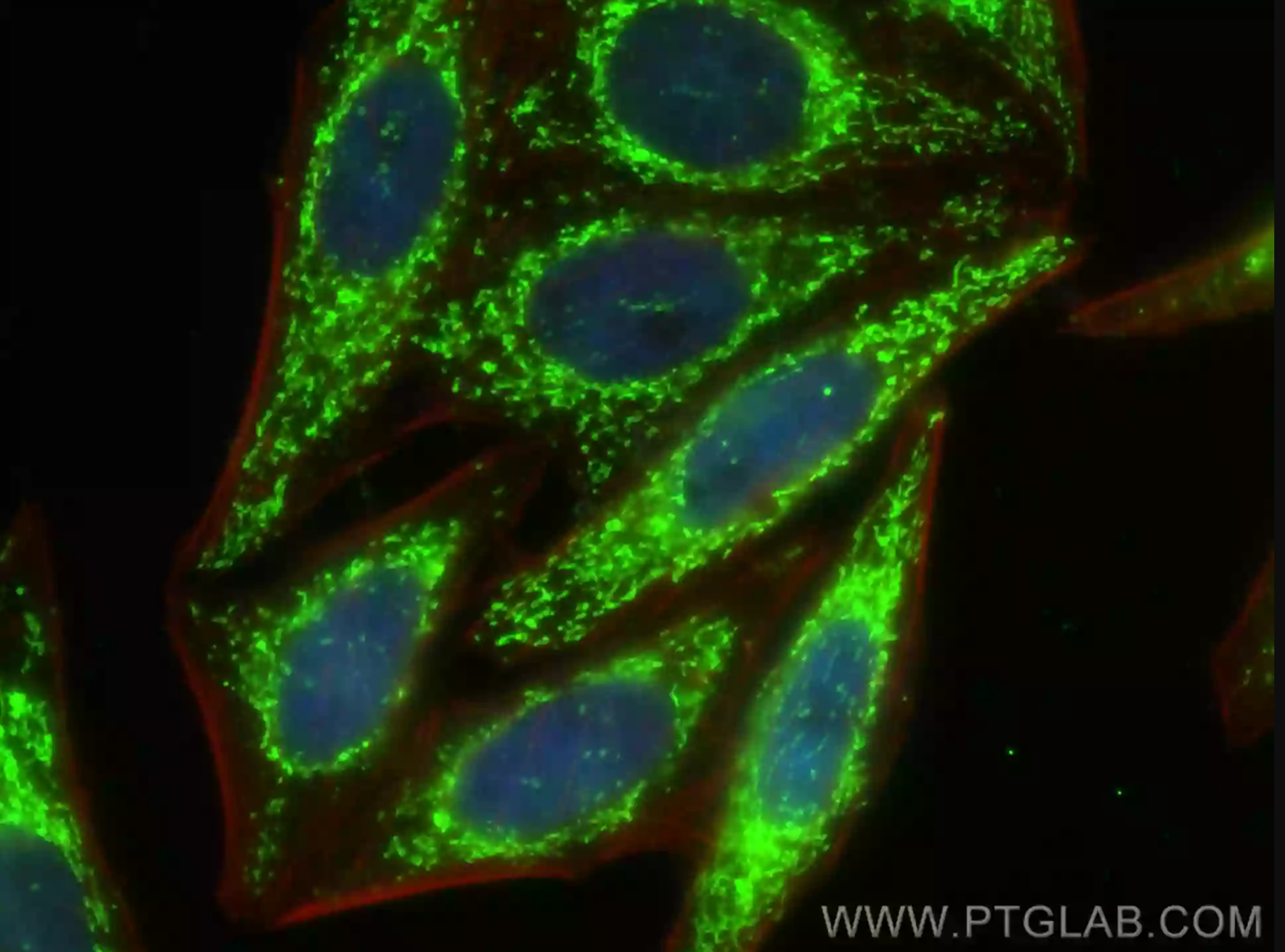
Featured Product LONP1 Polyclonal antibody (15440-1-AP)
Immunofluorescent analysis of (4% PFA) fixed HepG2 cells using LONP1 antibody (15440-1-AP) at dilution of 1:200 and CoraLite®-488-Conjugated AffiniPure Goat Anti-Rabbit IgG(H+L), CL594-Phalloidin (red). |
| COX5B Polyclonal antibody | (11418-2-AP) |
| DNAJA1 Polyclonal antibody | (11713-1-AP) |
| HSP70 Polyclonal antibody | (10995-1-AP) |
| LONP1 Polyclonal antibody | (15440-1-AP) |
View our range of mitochondrial antibodies and related products here, with our top-cited products listed:
| BAX Polyclonal antibody | (50599-2-Ig) |
| TOM20 Polyclonal antibody | (11802-1-AP) |
| COXIV Polyclonal antibody | (11242-1-AP) |
| SOD2 Polyclonal antibody | (24127-1-AP) |