ユビキチン化研究を効率的に進める方法:Ubiquitin-Trapの活用法とは?
タンパク質分解の鍵を握る「ユビキチン」の役割と多様な機能を解説するとともに、最新研究ツールの「Ubiquitin-Trap」がユビキチン研究の課題をどのように解決するかについて紹介します。
はじめに
ユビキチン(Ubiquitin)とは、主にタンパク質恒常性(プロテオスタシス)の制御因子として機能する、76残基のアミノ酸からなる分子量の小さなタンパク質です(分子量:約8.6 kDa)。ユビキチンをコードする遺伝子は生物種間で高度に保存されており、すべての真核生物の細胞内で恒常的・普遍的に発現します。ユビキチンは標的タンパク質に付加される翻訳後修飾の1つであり、「ユビキチン・プロテアソーム系(UPS)」と呼ばれるタンパク質分解の仕組みに深く関わることが知られています。また、構造的にユビキチンと類似したタンパク質(例:SUMO、NEDD8、ISG15他)は「ユビキチン様タンパク質(UBLs:ubiquitin-like proteins)」と呼ばれるタンパク質ファミリーを形成し、ユビキチンはユビキチン様タンパク質とともに翻訳後修飾タンパク質の大規模なファミリーを構成しています。ユビキチンはターゲットタンパク質を分解するために働くプロテアソーム依存的なタンパク質分解経路で中心的役割を担うだけではなく、近年ではオートファジー、細胞内シグナル伝達、エンドサイトーシス等の様々な役割を果たすことが明らかになっています。
ユビキチン化のプロセス
タンパク質のユビキチン化とは、標的となる基質タンパク質のアミノ酸配列中に存在するリシン(Lys、K)残基に対して特異的にユビキチンを転移させる一連の修飾プロセスを指します。ユビキチンと基質の結合は3段階の連続的な酵素反応によって進行し、その酵素カスケードは、ユビキチン活性化酵素(E1)、ユビキチン結合酵素(E2)、およびユビキチン転移酵素(E3、ユビキチンリガーゼ)の3つの異なる酵素が担います。はじめにユビキチンの活性化ステップでは、ユビキチン活性化酵素(E1)がATPの加水分解により生じたエネルギーを利用してユビキチン分子に結合します。続いて、ユビキチン活性化酵素(E1)の作用によってユビキチン結合酵素(E2)上にユビキチン分子が移動します。さらに、ユビキチン結合酵素(E2)-ユビキチン複合体はユビキチン転移酵素(E3)と会合し、ユビキチン転移酵素(E3)が基質となるタンパク質を認識するとともにユビキチン結合酵素(E2)から基質タンパク質中のリシン残基へのユビキチンの転移・結合が媒介されます。さらに、ユビキチンを構成するアミノ酸配列には7つのリシン残基が含まれるため、基質タンパク質に付加されたユビキチン中の7つのリシン残基(K6/Lys6、K11/Lys11、K27/Lys27、K29/Lys29、K33/Lys33、K48/Lys48、K63/Lys63)を介して、さらにユビキチン分子が付加・連結し、ユビキチン鎖を形成することが知られています。この現象は「ポリユビキチン化」と呼ばれます。基質タンパク質に結合するユビキチンは、モノユビキチン化とポリユビキチン化によってその形態が異なり、ポリユビキチン化においてもユビキチン分子の何番目のリシン残基にユビキチンが結合するのか、ユビキチン鎖の分岐や結合型の異なるユビキチン鎖が混合しているのか等によっても形態が異なります。なお、リシン残基を介したユビキチン同士の結合と比べると典型的な修飾型ではありませんが、ユビキチンのN末端のメチオニン(M1/Met1)残基を介して直鎖状のポリユビキチン鎖も生じます。
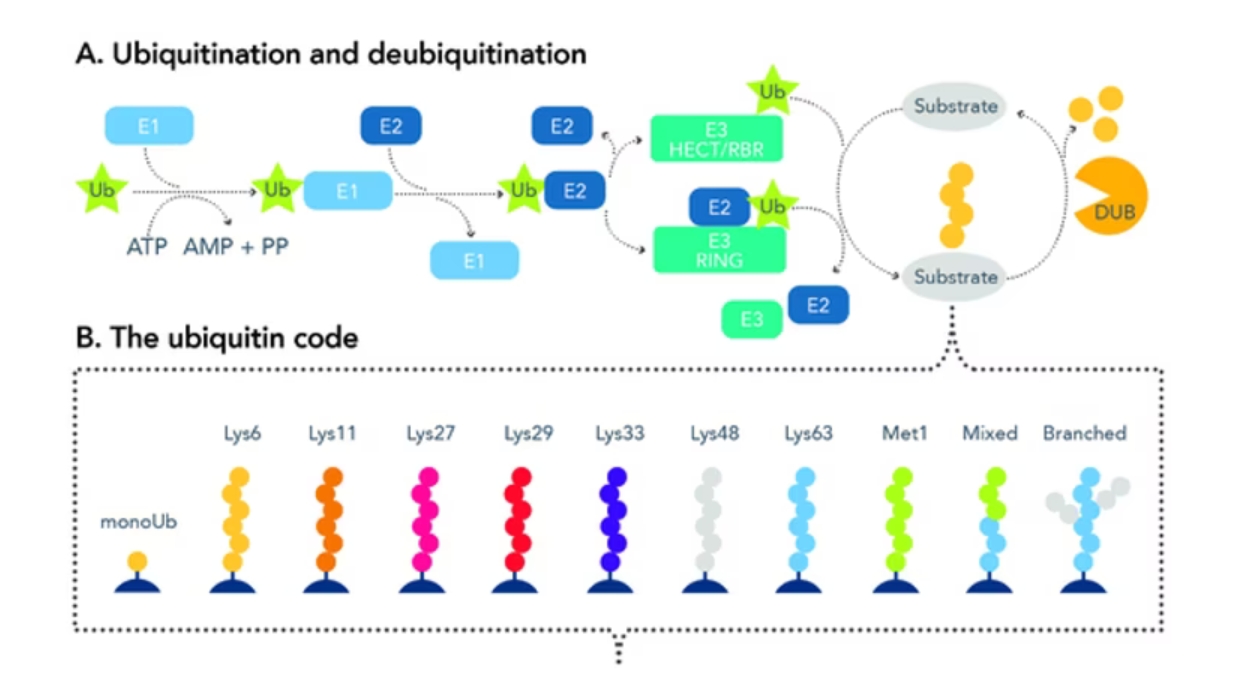
図1. ユビキチン化および脱ユビキチン化プロセスの概要。
ユビキチンコード
ユビキチン化が引き金となって発生する生物学的事象は、基質タンパク質がモノユビキチンとポリユビキチンのいずれの鎖長で修飾されるのか、ポリユビキチン化の場合、何番目のリシン残基にユビキチン鎖が結合するのか等のユビキチン結合型によって変化します。異なるユビキチンの結合型はそれぞれ固有のシグナル伝達機能や役割を発揮することで知られ、このことは「ユビキチンコード」とも称されます。例えば、モノユビキチン化は一般的にエンドサイトーシス、ヒストン修飾、DNA損傷応答等の非タンパク質分解性の事象を制御することが知られています。また、K48を介したポリユビキチン化は、ユビキチンが関与するシグナル伝達経路の中でも最も良く知られているユビキチン・プロテアソーム系のシグナルとして働き、プロテアソーム依存的なタンパク質分解を誘導します。文献で良く取り上げられるK48やK63以外の結合型は未解明な部分も多く存在しますが、ユビキチン化を介したシグナル伝達が関連して発生し得る生物学的事象の概要を以下の表に記載しました。
|
結合部位 |
ユビキチン鎖長 |
下流で発生する生物学的事象 |
|
基質特異的リシン残基 |
モノユビキチン化(単量体) |
エンドサイトーシス、ヒストン修飾、DNA損傷応答 |
|
K48 |
ポリユビキチン化(重合体) |
ユビキチン化タンパク質の分解 |
|
K63 |
ポリユビキチン化(重合体) |
免疫応答、炎症反応、リンパ球活性化 |
|
K6 |
ポリユビキチン化(重合体) |
抗ウイルス反応、オートファジー、マイトファジー、DNA修復 |
|
K11 |
ポリユビキチン化(重合体) |
細胞周期進行、プロテアソーム性タンパク質分解 |
|
K27 |
ポリユビキチン化(重合体) |
DNA複製、細胞増殖 |
|
K29 |
ポリユビキチン化(重合体) |
神経変性疾患、Wntシグナル伝達の下方制御、オートファジー |
|
M1 |
ポリユビキチン化(重合体) |
細胞死、免疫系に関連するシグナル伝達 |
このような典型的なユビキチン化の他に、ユビキチン鎖はアセチル化、リン酸化、分岐等を介してさらに修飾を受けます。また、ユビキチン鎖によるシグナル伝達とSUMOによるシグナル伝達が混在する場合もあります。
ユビキチン化研究の課題
ユビキチン(Ubiquitin)はその名が示す通りほぼ「普遍的(Ubiquitous)」に存在し、幅広い範囲の細胞機能に影響する制御因子ですが、研究者はユビキチン化のパターンやユビキチン化応答の特性解析を試みる中で様々な課題に直面しています。第一に、ユビキチンタンパク質は分子量が小さいため免疫原性が低いことが知られています。その結果、市販の多くのユビキチン抗体は非特異的反応を引き起こす懸念があり、多岐に及ぶアーチファクト物質との結合を生じるおそれがあります。また、ユビキチン化のプロセス自体もごく一過性で、脱ユビキチン化プロセスとともに可逆的な反応によって進行します。そのため、細胞ライセート中のユビキチン化タンパク質は極めて少量であることが多く、適切に検出するにはユビキチン化タンパク質の濃縮が必要です。さらに、基質選択性の異なる600種類以上のE3リガーゼが存在し、複数のリガーゼが同時に1つのタンパク質をユビキチン化する場合もあり、様々な結合パターンでタンパク質に付加されたユビキチンを検出できる試薬が必要となります。
ユビキチンおよびユビキチン化タンパク質を単離するクロモテックのUbiquitin-Trap
ユビキチン研究に伴う実験的課題を解決するために、クロモテック(2020年よりプロテインテックグループの一部になりました)は高親和性の「Nano-Trap(ナノトラップ)」シリーズとして、新たにユビキチンおよびユビキチン化タンパク質を単離するための「Ubiquitin-Trap(ユビキチントラップ)」を開発しました。Ubiquitin-Trapは、免疫沈降用のアガロースビーズまたは磁性アガロースビーズに結合した抗ユビキチンVHH抗体(別名:Nanobody®)からなる製品です。哺乳類、昆虫、植物、酵母等の幅広い生物由来の細胞ライセートから、単量体ユビキチン、ポリユビキチン鎖、ユビキチン化タンパク質を免疫沈降する際に使用できます。その他のNano-Trapシリーズの製品と同様にビーズ結合済みですぐに使用できる(Ready-to-use)状態で販売されており、迅速かつ簡便な目的タンパク質の免疫沈降/プルダウン実験に適用できます。VHH抗体は厳しい洗浄条件を適用しても安定性を示すため、夾雑物が少なくバックグラウンドの低い免疫沈降結果を得ることができます。
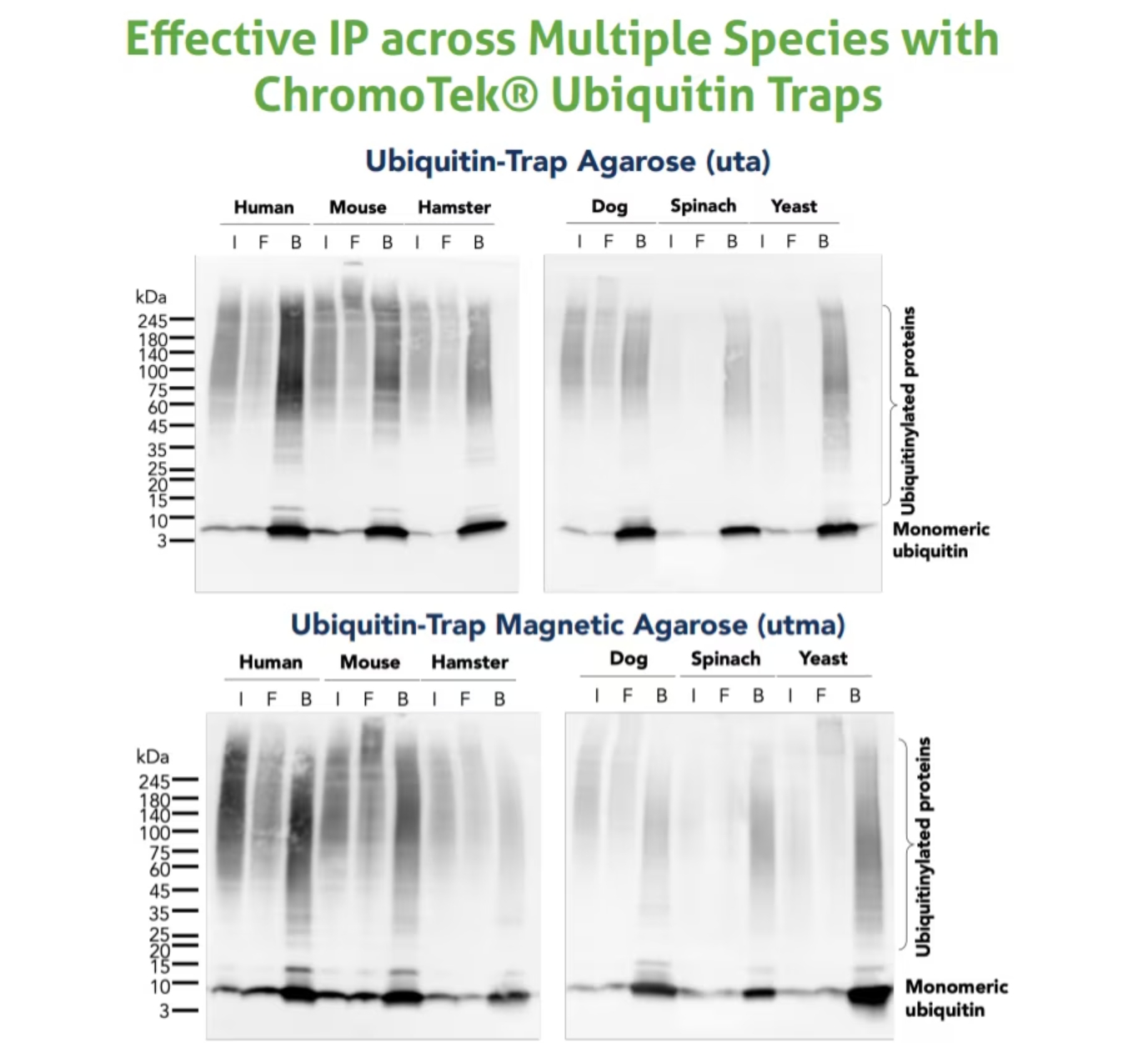
図2. Ubiquitin-Trap Agarose(カタログ番号:uta)とUbiquitin-Trap Magnetic Agarose(カタログ番号:utma)を使用した免疫沈降サンプルのウェスタンブロット(WB:Western blot)。試料:MG-132処理した、ヒト細胞(HEK293T)、マウス細胞(C2C12)、ハムスター細胞(CHO)、イヌ細胞(MDCK)、ホウレンソウ(Spinacia oleracea)、酵母(Saccharomyces cerevisiae)。各免疫沈降サンプルについて、インプットライセート(I)、フロースルー画分(F)、結合画分(B)をウェスタンブロットで解析しました。WBに使用した抗体:ユビキチン組換え抗体(カタログ番号:80992-1-RR)、HRP標識ヤギ抗ウサギIgG(H+L)抗体(カタログ番号:SA00001-2)。
免疫沈降用Ubiquitin-Trap製品一覧
|
カタログ番号 |
製品名 |
サイズ/構成 |
|
Ubiquitin-Trap Agarose |
10 rxns、20 rxns、100 rxns |
|
|
Ubiquitin-Trap Magnetic Agarose |
10 rxns、20 rxns、100 rxns |
|
|
Ubiquitin-Trap Agarose Kit |
20 rxns |
|
|
Ubiquitin-Trap Magnetic Agarose Kit |
20 rxns (キット付属試薬:免疫沈降用Lysis buffer、RIPA buffer、Dilution buffer、Wash buffer、Elution buffer) |
ウェスタンブロット用ユビキチン抗体および検出用二次抗体一覧
|
カタログ番号 |
製品名 |
交差種 |
|
Ubiquitin Polyclonal Antibody |
ヒト、マウス、ラット |
|
|
Ubiquitin Recombinant Antibody |
ヒト、マウス、ラット、ハムスター、酵母、イヌ、ホウレンソウ |
|
|
Multi-rAB HRP-Goat Anti-Rabbit Recombinant Secondary Antibody (H+L) |
ウサギ |
|
|
HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) |
ウサギ |
|
|
Biotin-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) |
ウサギ |
|
|
Alkaline Phosphatase-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) |
ウサギ |
FAQ
- SDS-PAGEやウェスタンブロットを実施すると、スメアなバンド(縦方向にブロード状のバンド)でユビキチンが観察されます。理由を教えてください。
- Ubiquitin-Trapはユビキチンモノマー、ポリユビキチン鎖、ユビキチン化タンパク質と結合します。そのため、免疫沈降後のビーズ結合画分は様々な鎖長および分子量のユビキチン化タンパク質を含有し、ゲルやブロット上にスメアなバンドが出現します。
- Ubiquitin-Trapは異なる結合型のユビキチン鎖を区別することができますか?
- Ubiquitin-Trapは特定の結合型のユビキチン特異的な製品ではありません。異なる形態・結合型の多様なユビキチン鎖と結合します。そのため、ユビキチン鎖の結合型を区別する必要がある場合は、ウェスタンブロットの際に目的とする結合型に特異的な抗体の使用を検討してください。
- Ubiquitin-Trapの結合能(結合キャパシティ)を教えてください。
- ユビキチンは様々な鎖長を形成するため、Ubiquitin-Trapの正確な結合能(結合キャパシティ)は完全に定義されていません。ユビキチン鎖は一か所に結合する場合や、複数の部位に結合する場合があるため、総合的な結合能の測定は容易ではなく、お示しできる数値はありません。
- Ubiquitin-Trapに使用されているユビキチンVHH抗体(別名:Nanobody®)の未標識体を単品購入できますか?
- 可能です。プロテインテックは、カスタム標識や抗体固相化実験に利用可能な未標識ユビキチンVHH抗体を販売しています。
- Ubiquitin-TrapはIP-MS(免疫沈降‐質量分析)のワークフローに適用できますか?
- 適用できます。その際、Ubiquitin-Trapはオンビーズ消化法に最適な製品であり、同手法を用いることを推奨します。オンビーズ消化法によるIP-MSを実施する場合は、プロテインテックのプロトコール「On-bead digest protocol for mass spectrometry」(PDF[言語:英語])をご参照ください。
- 細胞サンプル中のユビキチン化タンパク質の収量を増やし、ユビキチン化タンパク質を維持する方法を教えてください。
- 細胞サンプル中のユビキチン化タンパク質は、MG-132等のプロテアソーム阻害剤で処理することで保護・維持できます。最適な実験条件は、細胞の種類に応じて実験ごとに検討する必要があります。まずは細胞を5~25 µM MG-132で1~2時間インキュベーションすると良いでしょう。MG-132は過剰に暴露すると、細胞毒性を示すので注意してください。
参考文献


