Validation Data Gallery
Tested Applications
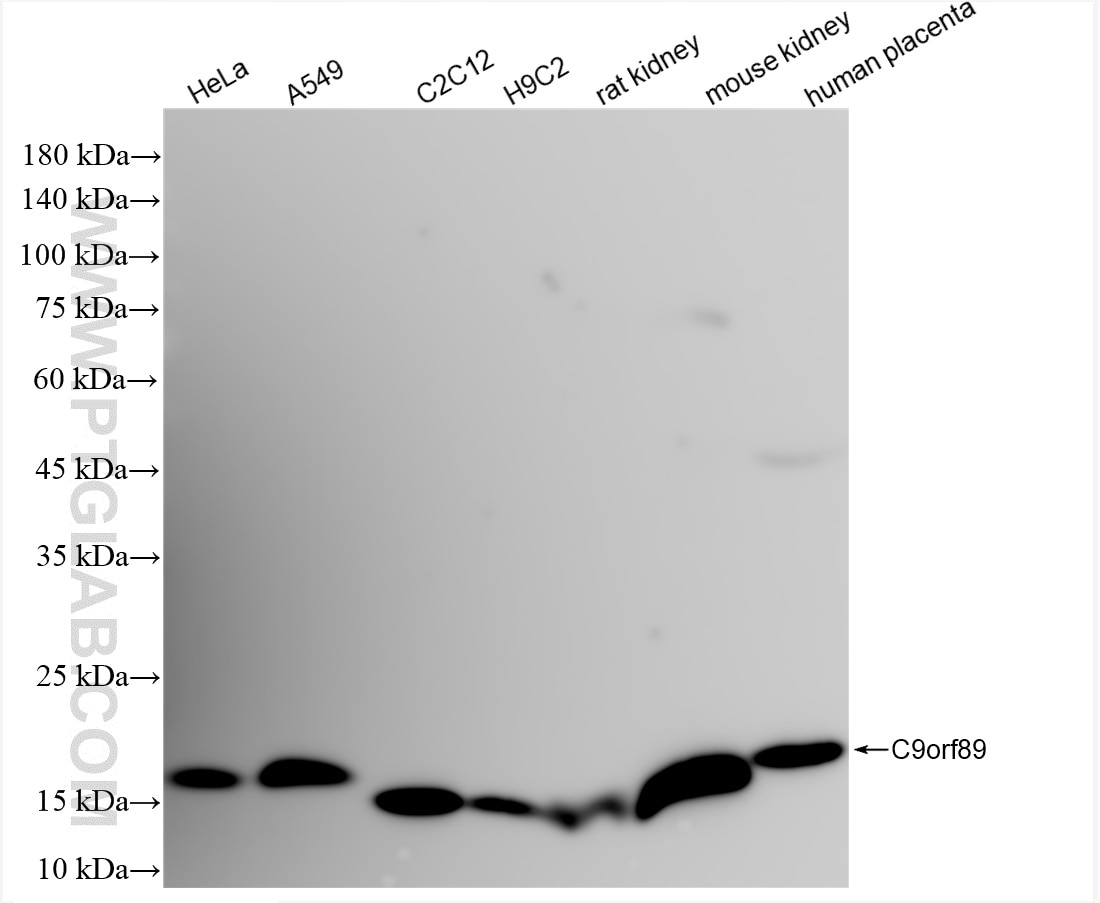
| Positive WB detected in | HeLa cells, A549 cells, C2C12 cells, H9C2 cells, rat kidney tissue, mouse kidney tissue, human placenta tissue |
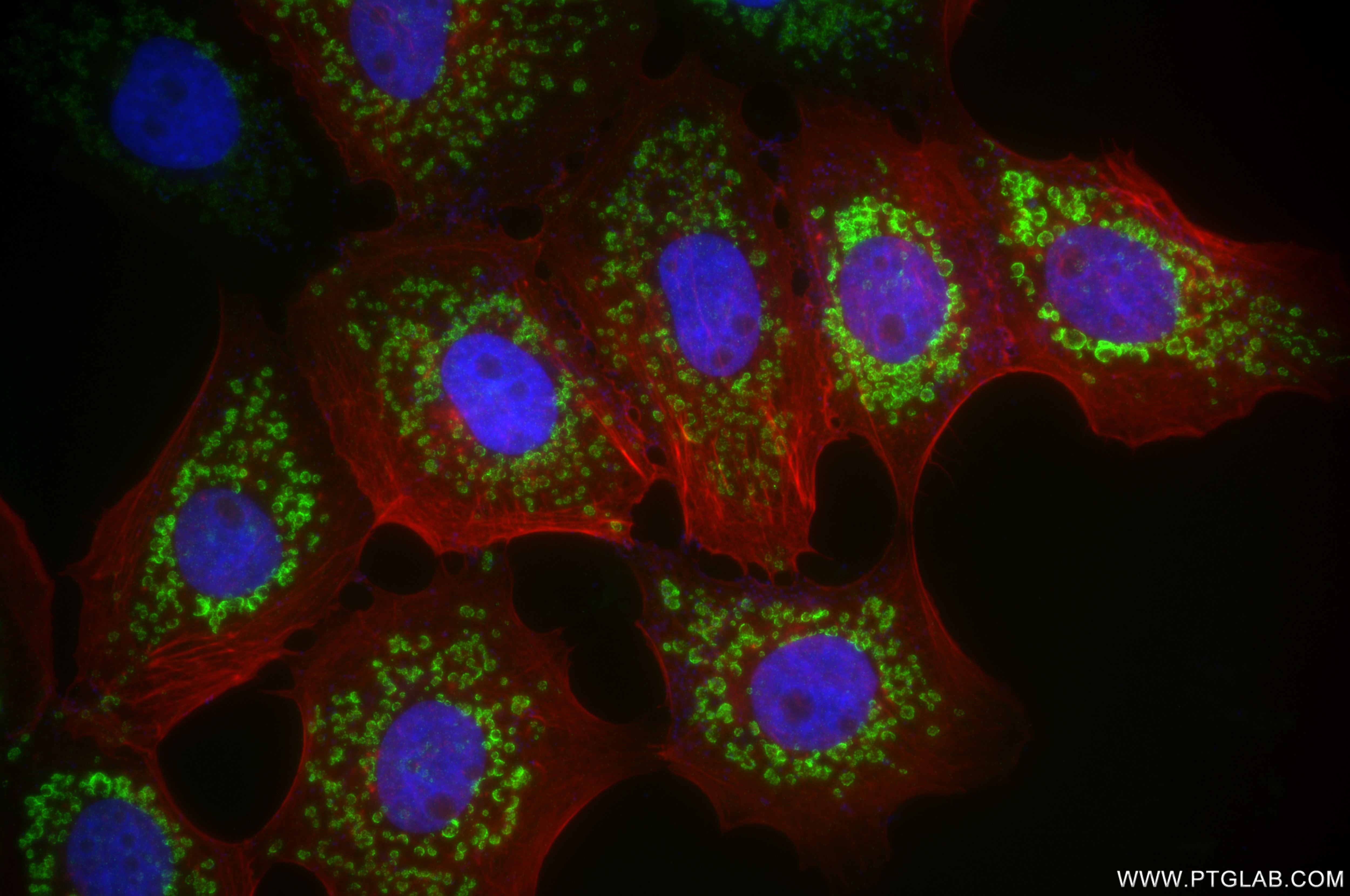
| Positive IF/ICC detected in | A431 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:250-1:1000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
85357-1-RR targets C9orf89 in WB, IF/ICC, ELISA applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | C9orf89 fusion protein Ag14084 相同性解析による交差性が予測される生物種 |
| Full Name | chromosome 9 open reading frame 89 |
| Calculated molecular weight | 228 aa, 26 kDa |
| Observed molecular weight | 15-21 kDa |
| GenBank accession number | BC004500 |
| Gene Symbol | C9orf89 |
| Gene ID (NCBI) | 84270 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | Q96LW7 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
C9orf89 (also known as CARD19 or BinCARD) is a mitochondria-localized protein that inhibits BCL10-induced NF-κB activation mainly through its caspase recruitment structural domain (CARD). It plays a role in T cell receptor (TCR) signaling, but deletion of endogenous CARD19 has a lesser effect on Bcl10-dependent NF-κB activation. In addition, C9orf89 is expressed in multiple cell types and is involved in the regulation of inflammatory and immune responses.
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for C9orf89 antibody 85357-1-RR | Download protocol |
| IF protocol for C9orf89 antibody 85357-1-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |