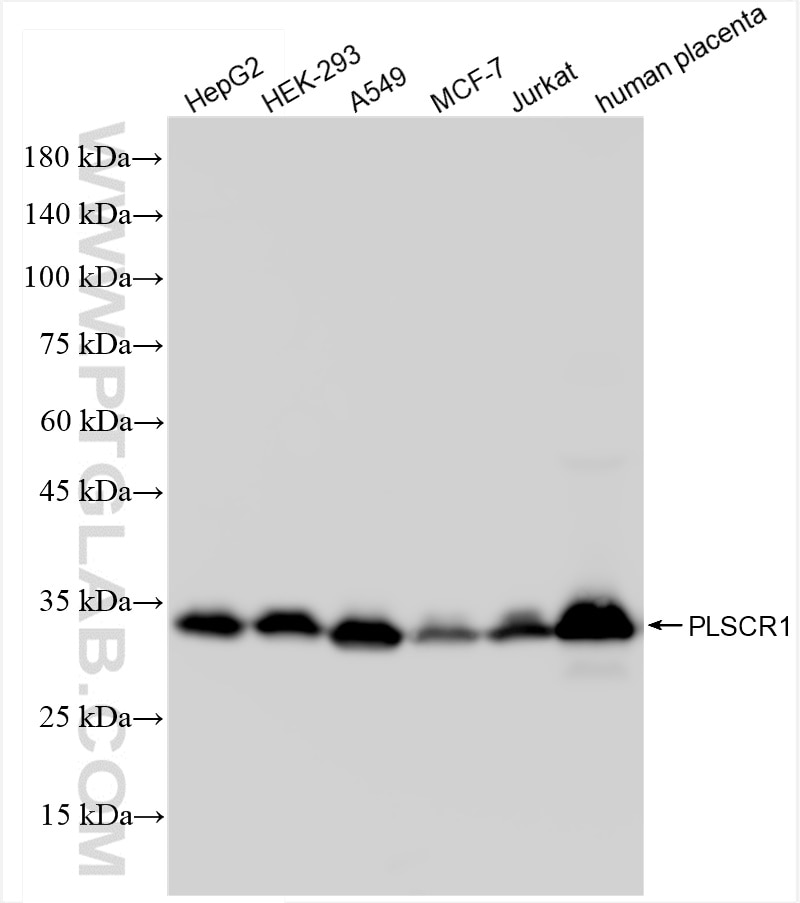
Validation Data Gallery
Tested Applications
| Positive WB detected in | HepG2 cells, HEK-293 cells, A549 cells, MCF-7 cells, Jurkat cells, human placenta tissue |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
84888-4-RR targets PLSCR1 in WB, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | PLSCR1 fusion protein Ag2172 相同性解析による交差性が予測される生物種 |
| Full Name | phospholipid scramblase 1 |
| Calculated molecular weight | 318 aa, 35 kDa |
| Observed molecular weight | 34 kDa |
| GenBank accession number | BC021100 |
| Gene Symbol | PLSCR1 |
| Gene ID (NCBI) | 5359 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purfication |
| UNIPROT ID | O15162 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
PLSCR1 (Phospholipid scramblase 1) is a member of the family of membrane proteins that mediate the transbilayer movement of phospholipids ("scrambling") in a Ca2+ dependent manner, which was identified as an a type II transmembrane protein involved in the calcium-dependent, nonspecific, rapid redistribution of phospholipids. A cysteine-rich palmitoylation motif [184CCCPCC189] of PLSCR1, a thioester linkage to the sulfhydryl groups of cysteines, controls the transportation of PLSCR1 to the cell membrane or nucleus (PMID: 32292520). PLSCR1 is strongly expressed in response to interferon (IFN) treatment and viral infection (PMID: 35138119, 23487022).
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for PLSCR1 antibody 84888-4-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |