Validation Data Gallery
Tested Applications
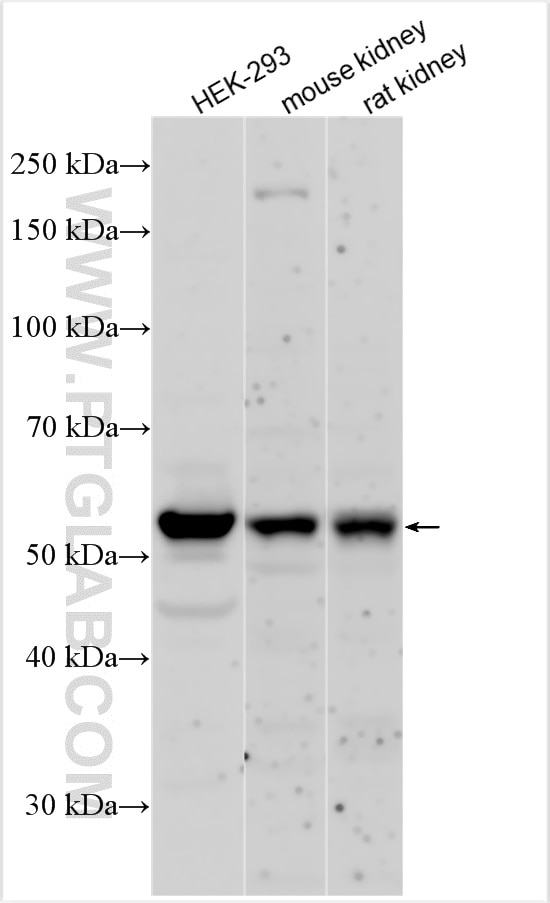
| Positive WB detected in | mouse kidney tissue, HEK-293 cells, rat kidney tissue |
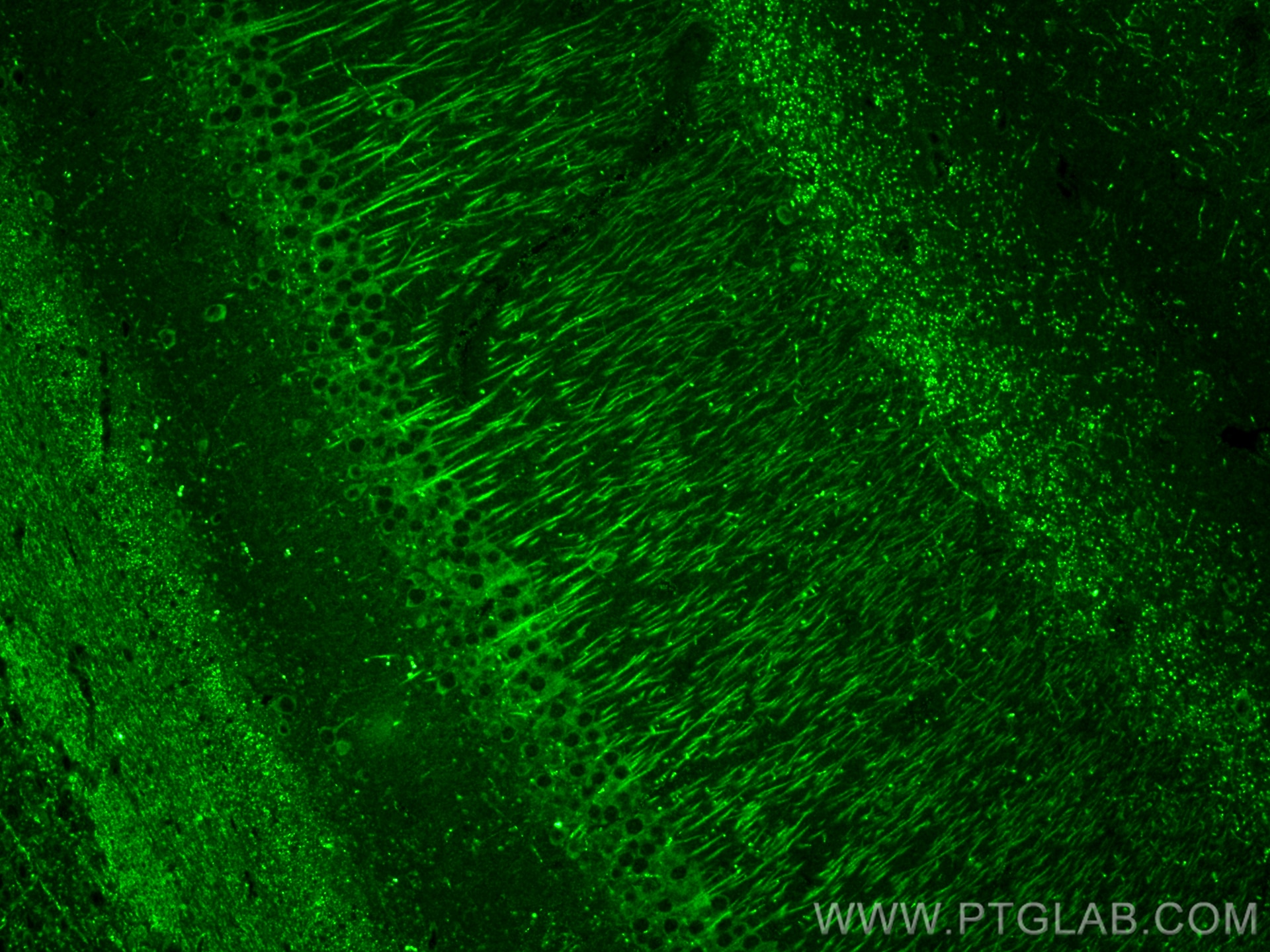
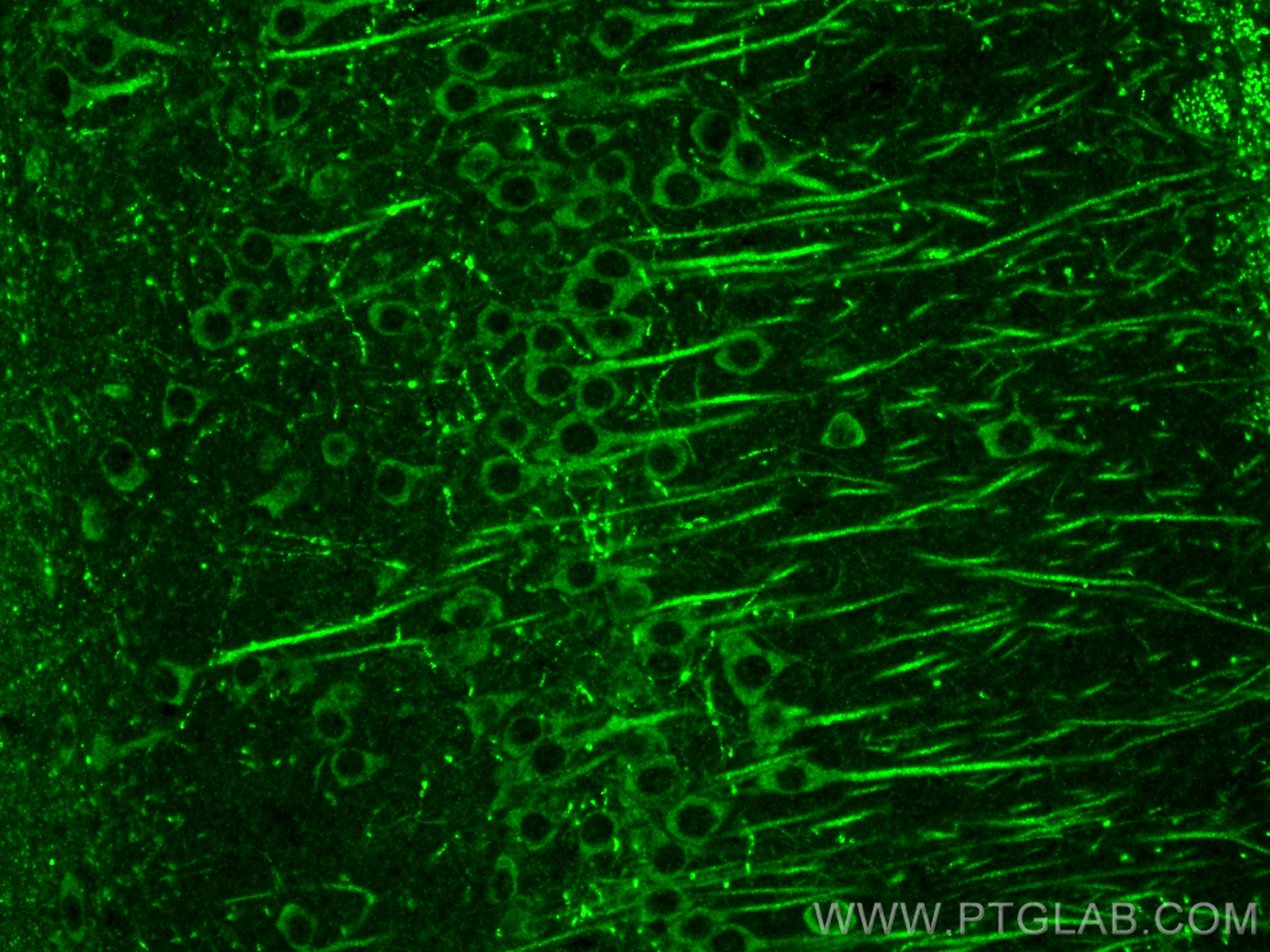
| Positive IF-P detected in | mouse brain tissue |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:500-1:1000 |
| Immunofluorescence (IF)-P | IF-P : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
30843-1-AP targets SLC6A20 in WB, IF-P, ELISA applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen | SLC6A20 fusion protein Ag33967 相同性解析による交差性が予測される生物種 |
| Full Name | solute carrier family 6 (proline IMINO transporter), member 20 |
| Observed molecular weight | 50-60 kDa |
| GenBank accession number | BC136431 |
| Gene Symbol | SLC6A20 |
| Gene ID (NCBI) | 54716 |
| RRID | AB_3669764 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity Purification |
| UNIPROT ID | Q9NP91 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
SLC6A20 also known as XT3, SIT1, belongs to the family of solute carrier 6, mediating the transport of neutral amino acids from the extracellular milieu toward cell or storage vesicles utilizing an electric membrane potential as the driving force. SLC6A20 is expressed in the kidney and small intestine and is localized in the brush marginal membrane of the proximal renal tubules (PMID: 16174864). Mutations in SLC6A20 are associated with Hyperglycinuria and Iminoglycinuria (PMID: 19033659)
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for SLC6A20 antibody 30843-1-AP | Download protocol |
| IF protocol for SLC6A20 antibody 30843-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |