Validation Data Gallery
Tested Applications
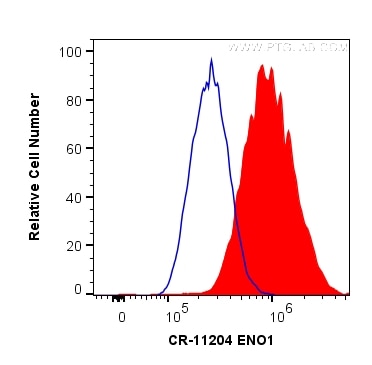
| Positive FC (Intra) detected in | HeLa cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Flow Cytometry (FC) (INTRA) | FC (INTRA) : 0.20 ug per 10^6 cells in a 100 µl suspension |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
CR-11204 targets ENO1 in FC (Intra) applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag1692 Product name: Recombinant human ENO1 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 2-434 aa of BC015641 Sequence: SILKIHAREIFDSRGNPTVEVDLFTSKGLFRAAVPSGASTGIYEALELRDNDKTRYMGKGVSKAVEHINKTIAPALVSKKLNVTEQEKIDKLMIEMDGTENKSKFGANAILGVSLAVCKAGAVEKGVPLYRHIADLAGNSEVILPVPAFNVINGGSHAGNKLAMQEFMILPVGAANFREAMRIGAEVYHNLKNVIKEKYGKDATNVGDEGGFAPNILENKEGLELLKTAIGKAGYTDKVVIGMDVAASEFFRSGKYDLDFKSPDDPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDDWGAWQKFTASAGIQVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLAQANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSKAKFAGRNFRNPLAK 相同性解析による交差性が予測される生物種 |
| Full Name | enolase 1, (alpha) |
| Calculated molecular weight | 47 kDa |
| Observed molecular weight | 47 kDa |
| GenBank accession number | BC015641 |
| Gene Symbol | ENO1 |
| Gene ID (NCBI) | 2023 |
| ENSEMBL Gene ID | ENSG00000074800 |
| RRID | AB_2934333 |
| Conjugate | Cardinal Red™ Fluorescent Dye |
| Excitation/Emission maxima wavelengths | 592 nm / 611 nm |
| 激发激光 | Yellow-Green Laser (561 nm) |
| Form | |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | P06733 |
| Storage Buffer | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
Enolase is an important glycolytic enzyme involved in the interconversion of 2-phosphoglycerate to phosphoenolpyruvate.Enolase were down-regulated in oxLDL-treated cells or in VLDL-treated cells .And it identified which are associated with glucose metabolism were down-regulated in the process of foam cells formation.(19756395)
Protocols
| Product Specific Protocols | |
|---|---|
| FC protocol for Cardinal Red™ ENO1 antibody CR-11204 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |