Validation Data Gallery
Filter:
Tested Applications
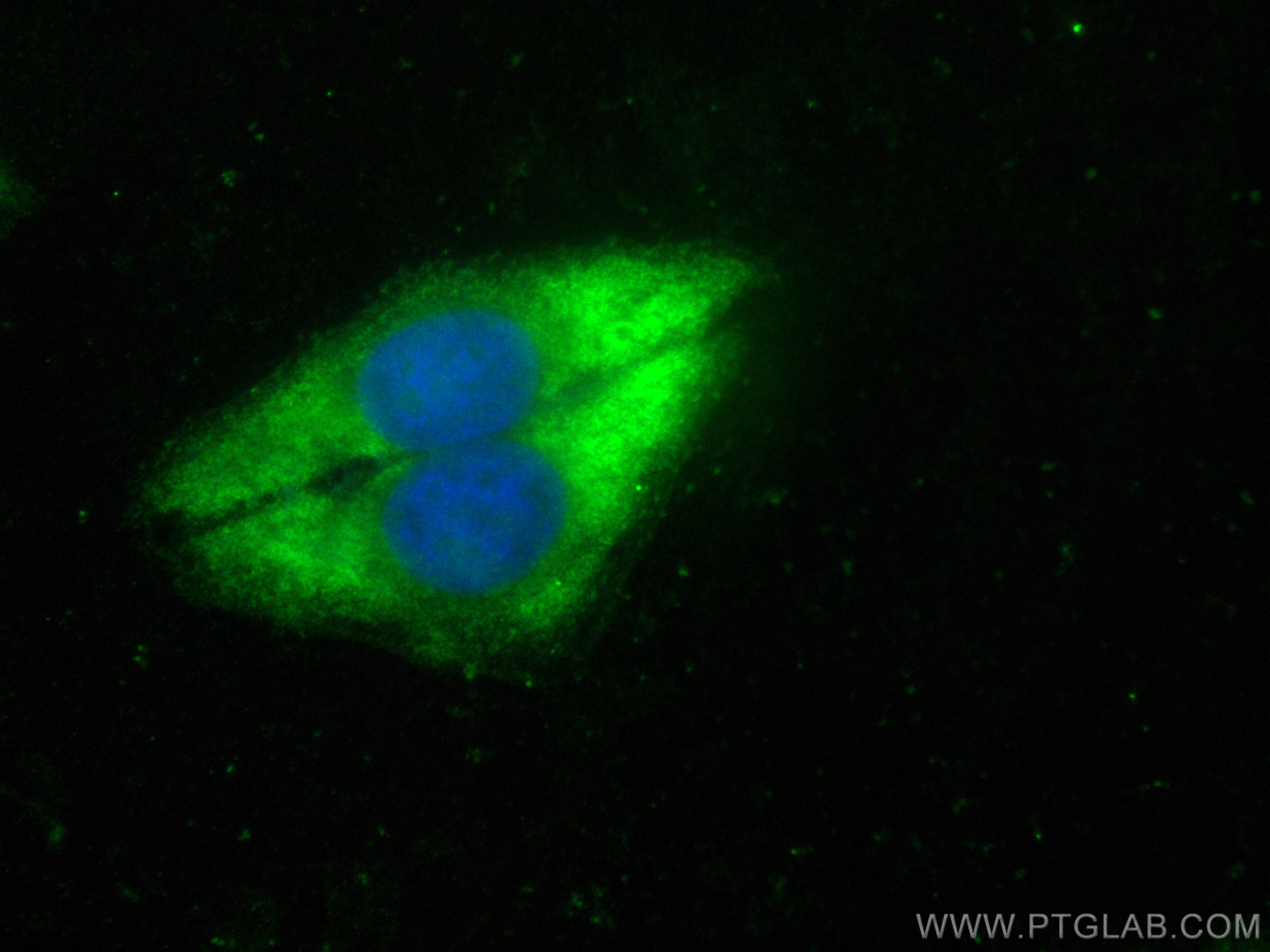
| Positive IF/ICC detected in | HepG2 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Published Applications
| WB | See 1 publications below |
Product Information
CL488-18295 targets HRAS in WB, IF/ICC applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Cited Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
Peptide 相同性解析による交差性が予測される生物種 |
| Full Name | v-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| Calculated molecular weight | 21 kDa |
| Observed molecular weight | 21 kDa |
| GenBank accession number | BC006499 |
| Gene Symbol | HRAS |
| Gene ID (NCBI) | 3265 |
| RRID | AB_3672694 |
| Conjugate | CoraLite® Plus 488 Fluorescent Dye |
| Excitation/Emission maxima wavelengths | 493 nm / 522 nm |
| 激发激光 | Blue laser (488 nm) |
| Form | |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | P01112 |
| Storage Buffer | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA{{ptg:BufferTemp}}7.3 |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
HRAS, also named as c-bas/has and RASH1, is v-Ha-ras Harvey rat sarcoma viral oncogene homolog from the mammalian ras gene family and it is a member of the small GTPase superfamily. RAS proteins are involved in signal transduction pathways, and bind GDP/GTP and possess intrinsic GTPase activity. Defects in HRAS are a cause of cancer. The antibody is specific to HRAS, can't recognize NRAS and KRAS.
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for CL Plus 488 HRAS antibody CL488-18295 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |