Validation Data Gallery
Tested Applications
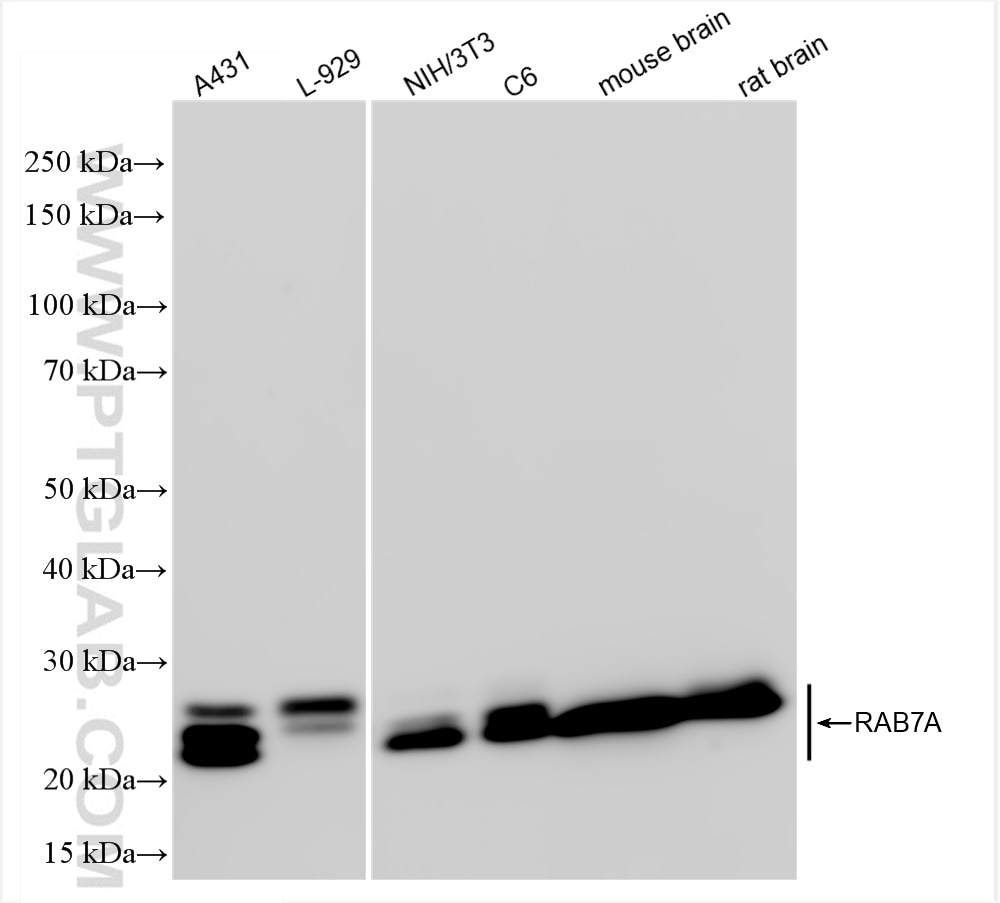
| Positive WB detected in | A431 cells, L-929 cells, NIH/3T3 cells, C6 cells, mouse brain tissue, rat brain tissue |
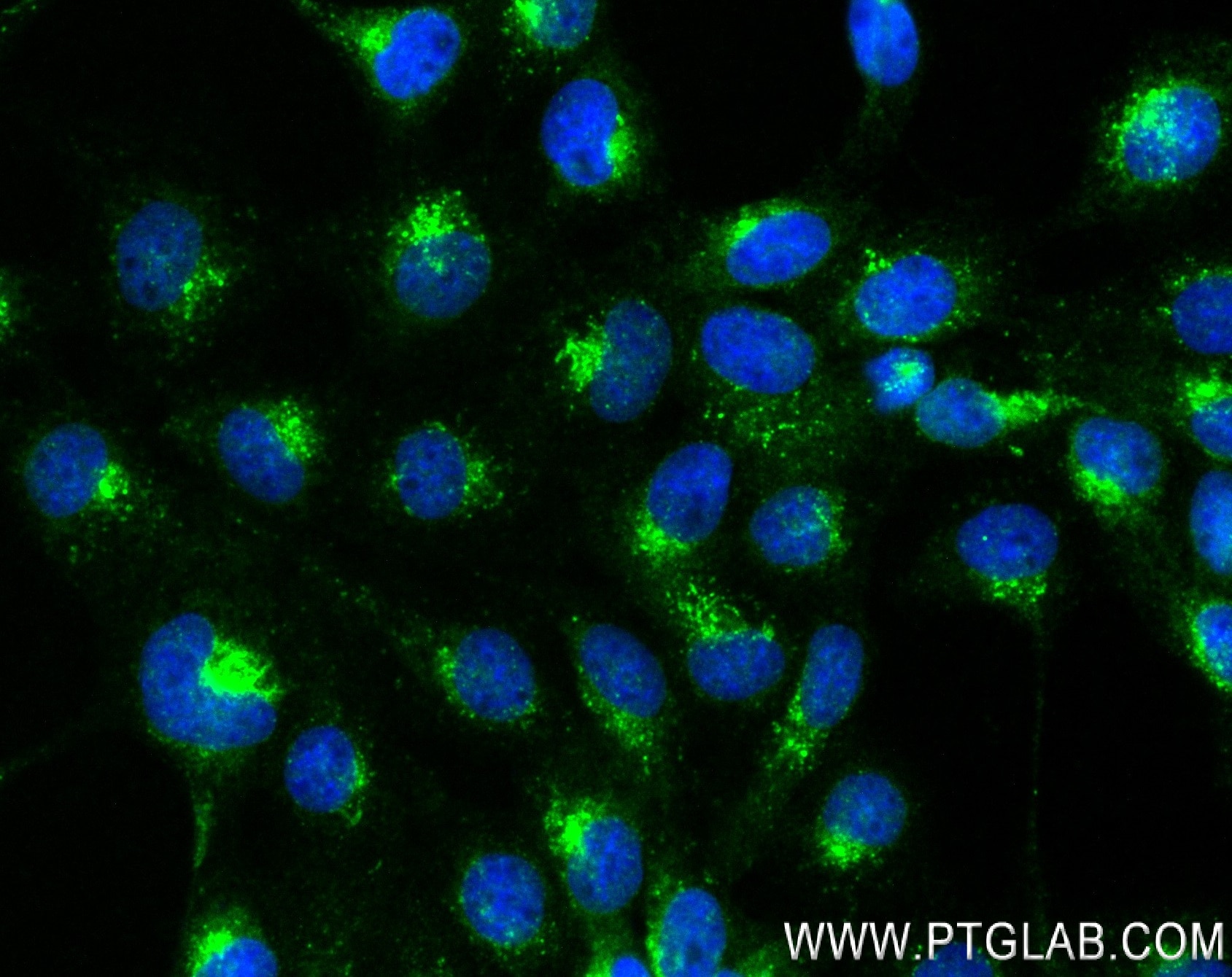
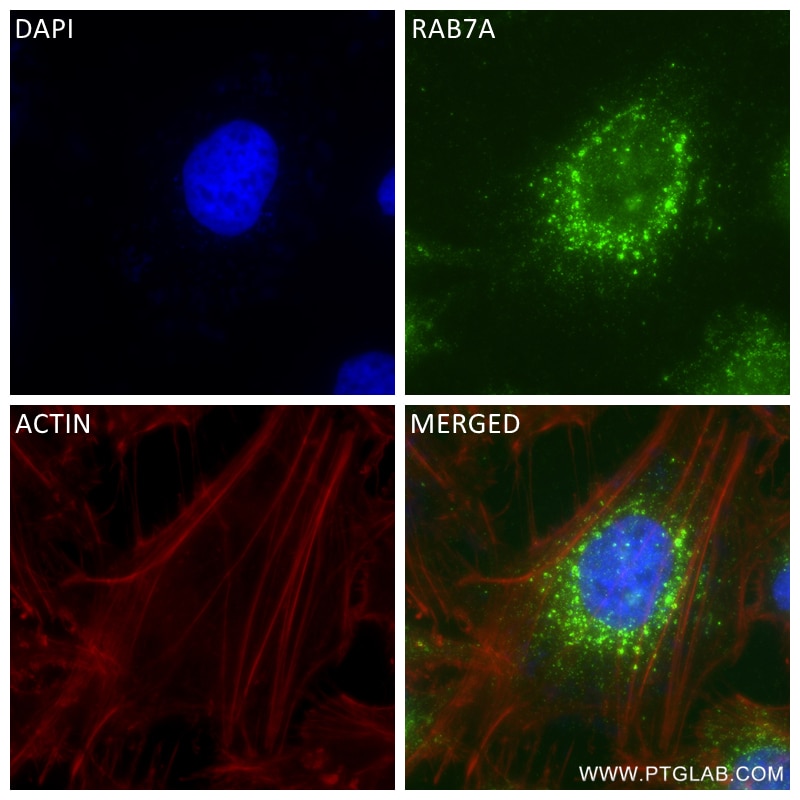
| Positive IF/ICC detected in | A431 cells, HeLa cells |
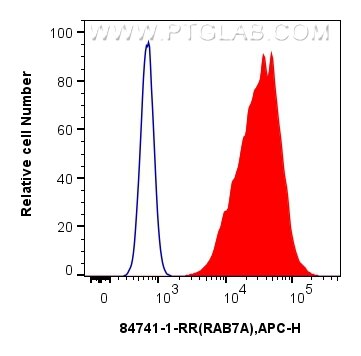
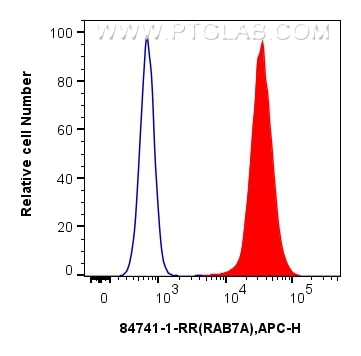
| Positive FC (Intra) detected in | K-562 cells, A431 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:200-1:800 |
| Flow Cytometry (FC) (INTRA) | FC (INTRA) : 0.25 ug per 10^6 cells in a 100 µl suspension |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
84741-1-RR targets RAB7A in WB, IF/ICC, FC (Intra), ELISA applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | Peptide 相同性解析による交差性が予測される生物種 |
| Full Name | RAB7A, member RAS oncogene family |
| Calculated molecular weight | 23 kDa |
| Observed molecular weight | 23 kDa |
| GenBank accession number | NM_004637 |
| Gene Symbol | RAB7A |
| Gene ID (NCBI) | 7879 |
| RRID | AB_3672126 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purfication |
| UNIPROT ID | P51149 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
RAB7A, also named as RAB7, belongs to the small GTPase superfamily and Rab family. It is involved in late endocytic transport. RAB7A contributes to the maturation of phagosomes (acidification). Defects in RAB7A are the cause of Charcot-Marie-Tooth disease type 2B (CMT2B). This antibody is specific to RAB7A.
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for RAB7A antibody 84741-1-RR | Download protocol |
| IF protocol for RAB7A antibody 84741-1-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |