Validation Data Gallery
Product Information
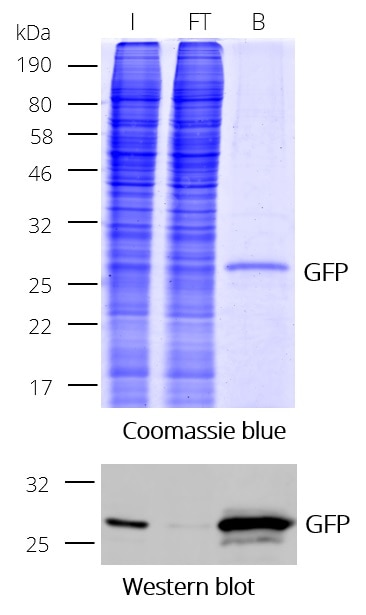
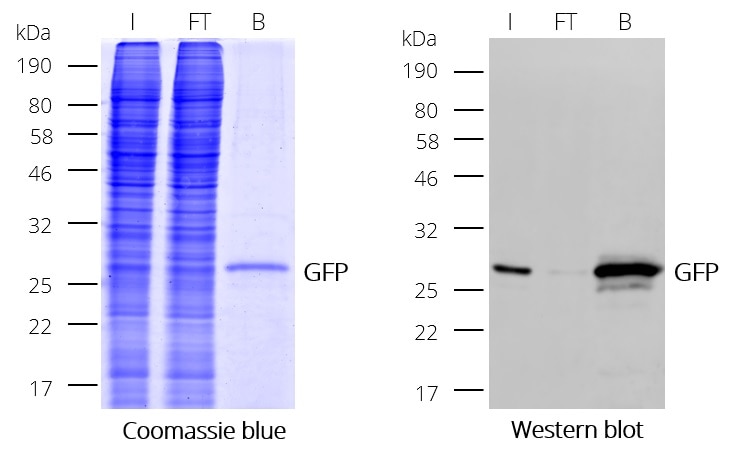
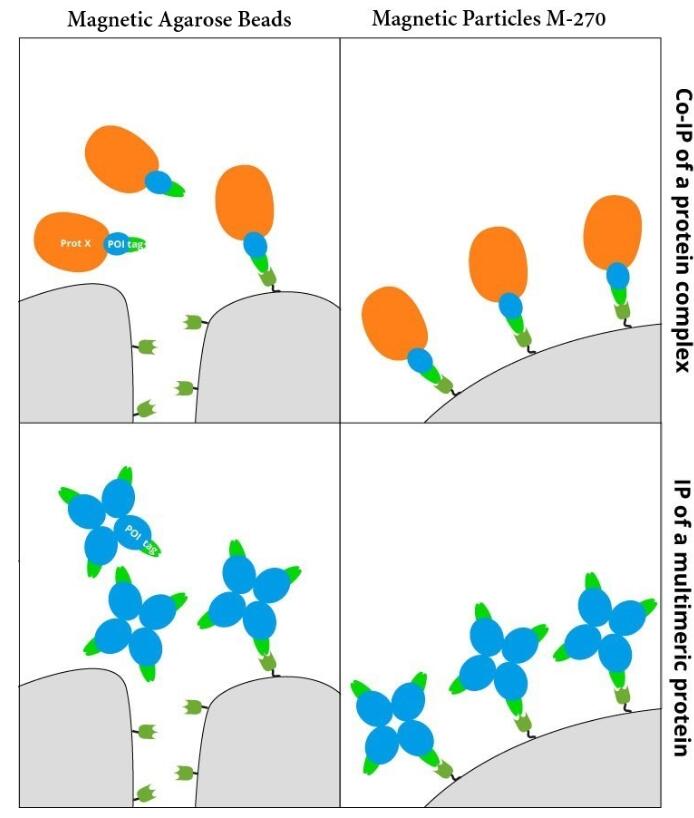
GFP-Trap® Magnetic Particles M-270 for immunoprecipitation (IP) of GFP-tagged proteins. GFP-Trap® Magnetic Particles M-270 is highly recommended, when very large proteins/complexes are investigated, and magnetic separation is needed for IP. It consists of a GFP VHH/ Nanobody coupled to Magnetic Particles M-270.
| Description | Immunoprecipitation of GFP-fusion proteins and their interacting factors with anti-GFP Nanobody conjugated to magnetic particles. GFP-Trap Magnetic Particles M-270 is recommended when very large proteins/complexes are investigated and magnetic separation is needed for IP.
• IP and Co-IP of GFP-tagged proteins without size limitation • Magnetic separation with easy and efficient washing of GFP-Trap Magnetic Particles M-270 • Automation and high throughput applications • No heavy & light antibody chains, short incubation (5-30 min) • Extraordinary binding, also under harsh conditions • Very high affinity (KD=1 pM) to bind even low abundant proteins |
| Applications | IP, CoIP, ChIP, RIP |
| Specificity/Target | AcGFP, Clover, eGFP, Emerald, GFP, GFP5, GFP Envy, GFP S65T, mGFP, mPhluorin, PA-GFP, Superfolder GFP, TagGFP, TagGFP2, monomeric eGFP A206K, CFP, YFP, Citrine, eCitrine, eYFP, Venus, Ypet, BFP For the complete list, please click here: Fluorescent protein specificity table |
| Binding capacity | 2-2.8 μg of recombinant GFP per 25 μL bead slurry |
| Conjugate | Magnetic Particles M-270, size: 2.8 µm high throughput-compatible |
| Elution buffer | SDS sample buffer 0.2 M glycine pH 2.5 |
| Wash buffer compatibility | 10 mM DTT, 8 M Urea, 2 M NaCl, 2 % Nonidet P40 Substitute, 0.2 % SDS, 1 % Triton X-100 |
| Type | Nanobody |
| Class | Recombinant |
| Host | Alpaca |
| Affinity (KD) | Dissociation constant KD of 1 pM |
| Compatibility with mass spectrometry | The GFP-Trap® is optimized for on-bead digestion. For the application note, please click here: On-bead digest protocol for mass spectrometry |
| RRID | AB_2827592 |
| Storage Buffer | PBS with 0.09% sodium azide |
| Storage Condition | Shipped at ambient temperature. Upon receipt store at 4°C. Stable for one year. Do not freeze! |
| Size | 25ul/reactions (eg:20rxns=500ul slurry) |
Documentation
| SDS |
|---|
| gtd_SDS_GFP-Trap® Magnetic Particles M-270 (EN) |
| Datasheet |
|---|
| GFP-Trap® Magnetic Particles M-270 Datasheet |
| GFP-Trap Specificity |
|---|
| Fluorescent protein specificity table |
| Trouble shooting |
|---|
| Troubleshooting guide immunoprecipitation (IP) |
Publications
| Application | Title |
|---|---|
Science DNSN-1 recruits GINS for CMG helicase assembly during DNA replication initiation in Caenorhabditis elegans | |
Nat Cell Biol ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16INK4a levels. | |
Mol Cell TNIP1 inhibits selective autophagy via bipartite interaction with LC3/GABARAP and TAX1BP1 | |
Mol Cell Redundant pathways for removal of defective RNA polymerase II complexes at a promoter-proximal pause checkpoint | |
Neuron Brain-derived autophagosome profiling reveals the engulfment of nucleoid-enriched mitochondrial fragments by basal autophagy in neurons. | |
Nat Commun Spatial control of avidity regulates initiation and progression of selective autophagy. |